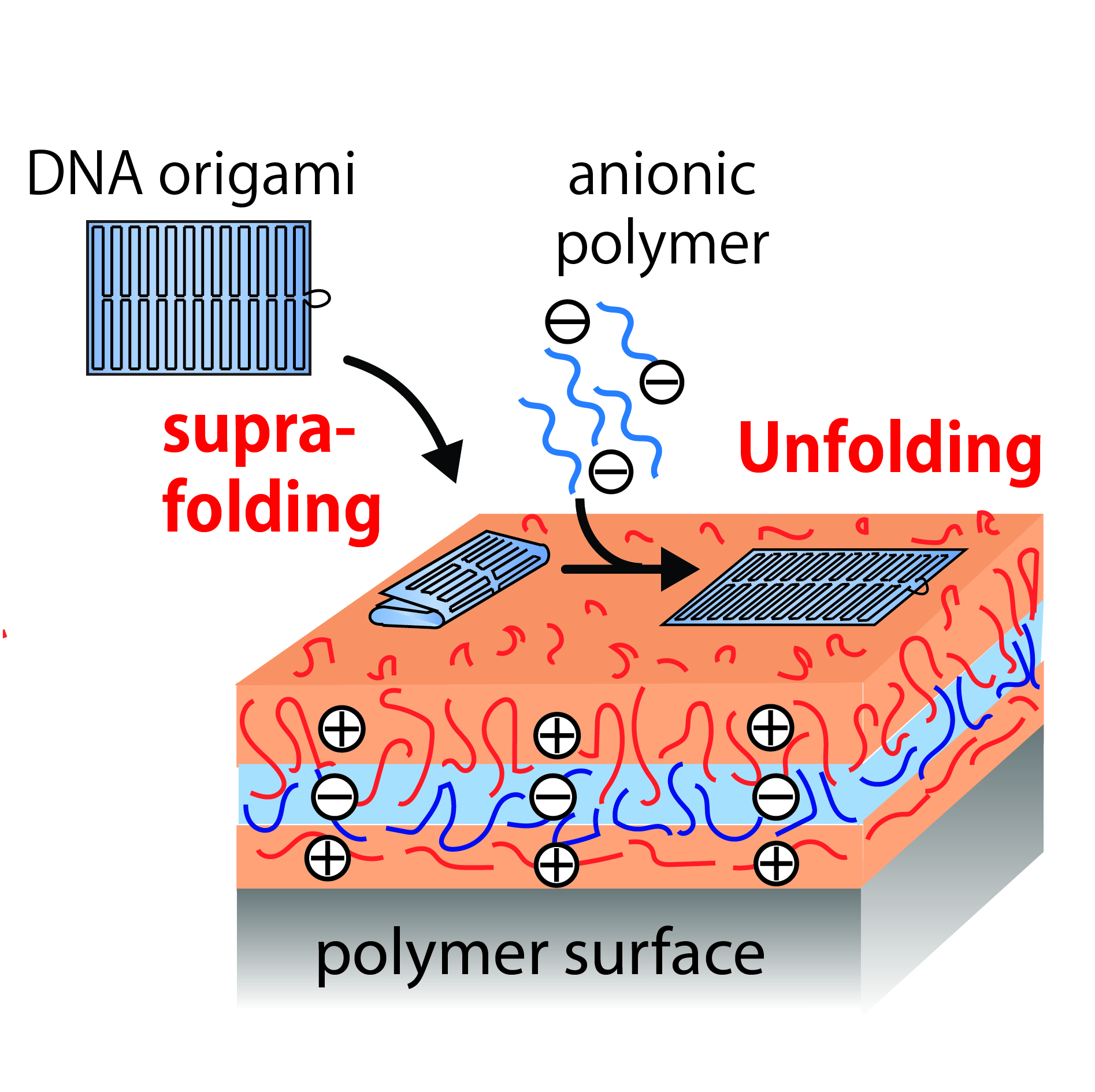
Abstract: We report that user-defined DNA nanostructures, such as two-dimensional (2D) origamis and nanogrids, undergo a rapid
higher-order folding transition, referred to as supra-folding, into three-dimensional (3D) compact structures (origamis) or well-defined
μm-long ribbons (nanogrids), when they adsorb on a soft cationic substrate prepared by layer-by-layer deposition of polyelectrolytes.
Once supra-folded, origamis can be switched back on the surface into their 2D original shape through addition of heparin, a highly
charged anionic polyelectrolyte known as an efficient competitor of DNA-polyelectrolyte complexation. Orthogonal to DNA base-pairing
principles, this reversible structural reconfiguration is also versatile; we show in particular that i) it is compatible with various origami
shapes, ii) it perfectly preserves fine structural details as well as sitespecific functionality, and iii) it can be applied to dynamically address
the spatial distribution of origami-tethered proteins.
Reference:
Reversible supra-folding of user-programmed functional DNA nanostructures on fuzzy cationic substrates
K. Nakazawa, F. El Fakih, V. Jallet, C. Rossi–Gendron, M. Mariconti, L. Chocron, M. Hishida, K. Saito, M. Morel, S. Rudiuk,* D. Baigl*
Angew. Chem. Int. Ed. 2021, 60, 15214 –15219
 -doi : 10.1002/anie.202101909
-doi : 10.1002/anie.202101909
Reference:
Reversible supra-folding of user-programmed functional DNA nanostructures on fuzzy cationic substrates
K. Nakazawa, F. El Fakih, V. Jallet, C. Rossi–Gendron, M. Mariconti, L. Chocron, M. Hishida, K. Saito, M. Morel, S. Rudiuk,* D. Baigl*
Angew. Chem. Int. Ed. 2021, 60, 15214 –15219